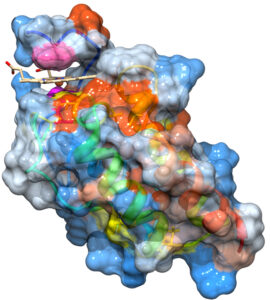
Csa2 crystal structure with the heme (stick model) positioned in the cleft between the CFEM domain and the N-terminal lobe
Due to the scarcity of free iron in the host environment, pathogenic microorganisms have by necessity evolved mechanisms to extract iron from host proteins. The heme cofactor of hemoglobin constitutes the largest iron pool in the animal body. C. albicans is able to extract heme from hemoglobin and to transport it into the cell, where is serves as a source of iron. We identified a family of extracellular proteins, the CFEM proteins, that capture heme from hemoglobin, shuttle it across the cell envelope and deliver it to the endocytic pathway. Our current research focuses on the mechanism of heme capture and transfer by CFEM proteins, and on the characterization of the transfer of heme from the CFEM network to the endocytic pathway.
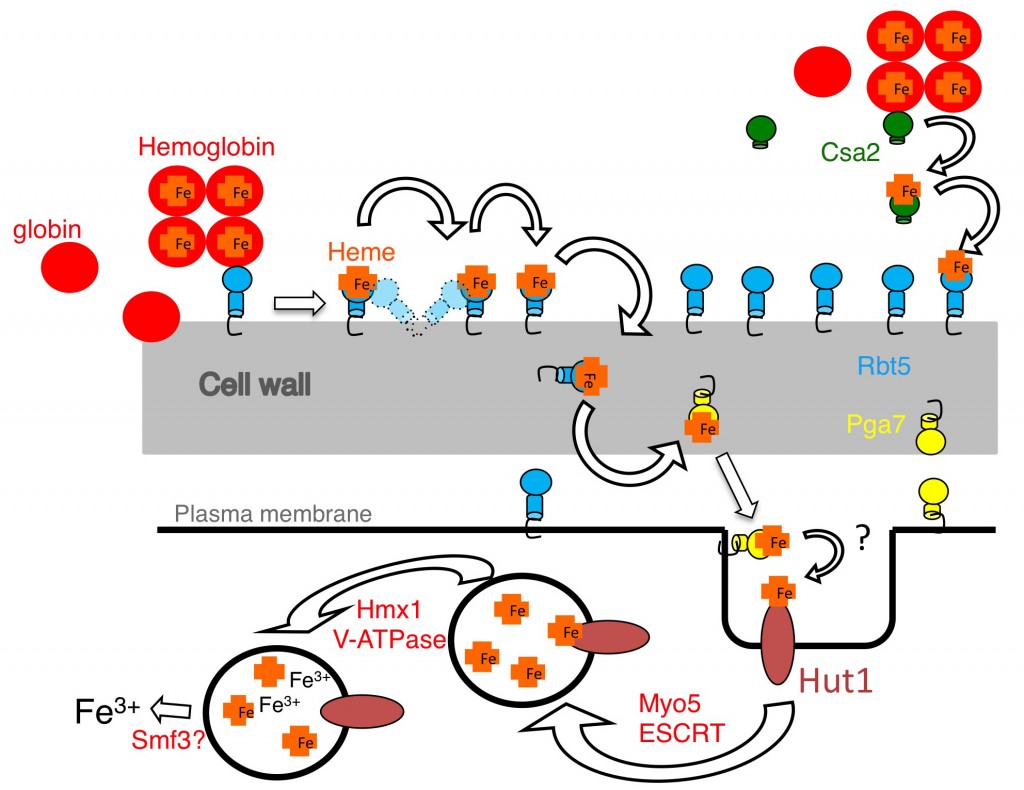
Our current understanding of the CFEM network of heme-iron acquisition is summarized in the model below. The C. albicans CFEM proteins Csa2 (a secreted hemophore), and Rbt5 and Pga7 (cell wall- and cell membrane-anchored hemophores) are able to extract heme from hemoglobin and to transfer it among themselves, from one protein to the next, across the cell wall. Heme is then taken up into the cell in an endocytosis-mediated pathway.
We are currently focusing on two aspects of the heme acquisition pathway:
a) Structure-function analysis of the CFEM proteins to understand the heme extraction and transfer reactions. This analysis is aided by the recent determination, in collaboration with Dr. Hay Dvir, of the crystal structure of the secreted CFEM protein Csa2.
Site-directed mutagenesis is used to modify candidate positions in the protein and to analyse the effect of these mutations on heme binding, extraction and transfer.
b) Identification of new factors involved in the heme utilization pathway. To do this, we implement genetic screens using C. albicans mutant collections such as the GRACE collection, as well as bioinformatic methods such as phylogenetic profiling. Several novel proteins involved in heme-iron utilization were isolated in this way and are currently being characterized.